Modeling the Early Stages of Phase Separation in Disordered Elastin-like Proteins
Abstract
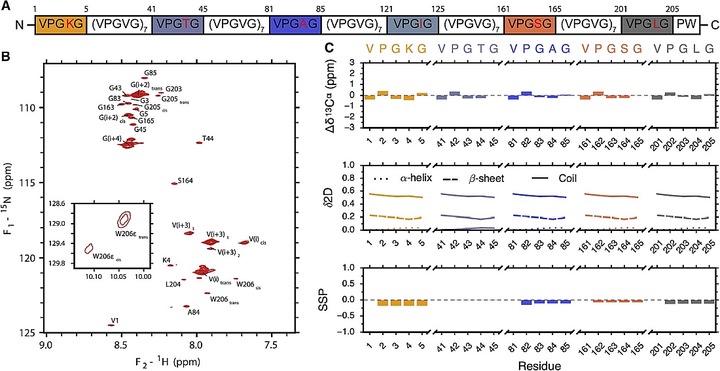
Elastin-like proteins (ELPs) are known to undergo liquid-liquid phase separation reversibly above a concentration-dependent transition temperature. Previous studies suggested that, as temperature increases, ELPs experience an increased propensity for type II β-turns. However, how the ELPs behave below the phase transition temperature itself is still elusive. Here, we investigate the importance of β-turn formation during the early stages of ELP self-association. We examined the behavior of two ELPs, a 150-repeat construct that had been investigated previously (ELP[V5G3A2-150] as well as a new 40-repeat construct (ELP40) suitable for nuclear magnetic resonance measurements. Structural analysis of ELP40 reveals a disordered conformation, and chemical shifts throughout the sequence are insensitive to changes in temperature over 20°C. However, a low population of β-turn conformation cannot be ruled out based on chemical shifts alone. To examine the structural consequences of β-turns in ELPs, a series of structural ensembles of ELP[V5G3A2-150] were generated, incorporating differing amounts of β-turn bias throughout the chain. To mimic the early stages of the phase change, two monomers were paired, assuming preferential interaction at β-turn regions. This approach was justified by the observation that buried hydrophobic turns are commonly observed to interact in the Protein Data Bank. After dimerization, the ensemble-averaged hydrodynamic properties were calculated for each degree of β-turn bias, and the results were compared with analytical ultracentrifugation experiments at various temperatures. We find that the temperature dependence of the sedimentation coefficient (so20,w) can be reproduced by increasing the β-turn content in the structural ensemble. This analysis allows us to estimate the presence of β-turns and weak associations under experimental conditions. Because disordered proteins frequently exhibit weak biases in secondary structure propensity, these experimentally-driven ensemble calculations may complement existing methods for modeling disordered proteins generally.
Supporting information can be found here.